MATH 427: ROC Curve and AUC
Announcements
On March 5th at 10am in JAAC, Kyle Mayer will be guest lecturing to talk about his role as a data analyst at Micron. Kyle has a Bachelors in Engineering and a Masters in Physics.
Bio: Kyle Mayer is a Senior Data Analytics Engineer at Micron Technology with over 15 years of experience. He currently supports the Global Quality organization with engineering and data science projects. Kyle specializes in modeling complex physical processes, delivering insights that drive business value. Leveraging his expertise in semiconductor manufacturing, Kyle has developed tools and processes that have prevented over $100M in lost revenue. His passion for applying artificial intelligence to solve complex problems and develop automated systems highlights his commitment to innovation. In his spare time, Kyle enjoys spending time with his kids and going on hikes.
Job Application 1
- Job Ad
- Directions and Resources
- Rubic (in progress)
- Due Next Friday: CV and Cover Letter
- Due Friday March 21st: whole application
Computational Set-Up
Default Dataset
A simulated data set containing information on ten thousand customers. The aim here is to predict which customers will default on their credit card debt.
Split the data
K-Nearest Neighbors Classifier: Build Model
- Response (\(Y\)):
default - Predictor (\(X\)):
balance
K-Nearest Neighbors Classifier: Predictions
Fitting a logistic regression
Fitting a logistic regression model with default as the response and balance as the predictor:
Making predictions in R
Binary Classifiers
- Start with binary classification scenarios
- With binary classification, designate one category as “Success/Positive” and the other as “Failure/Negative”
- If relevant to your problem: “Positive” should be the thing you’re trying to predict/care more about
- Note: “Positive” \(\neq\) “Good”
- For
default: “Yes” is Positive
- Some metrics weight “Positives” more and viceversa
Last Time
- Confusion Matrix
- Metrics based on confusion matrix
- Accuracy
- Recall/Sensitivity
- Precision/PPV
- Specificity
- NPV
- MCC
- F-Measure
- Today: ROC and AUC
Thresholding
Using a threshold
- Step 1: Predict probabilities for all observations
default_test_wprobs <- default_test |>
mutate(
knn_probs = predict(knnfit, new_data = default_test, type = "prob") |> pull(.pred_Yes),
logistic_probs = predict(logregfit, new_data = default_test, type = "prob") |> pull(.pred_Yes)
)
default_test_wprobs |> head() |> kable() # obtain probability predictions| default | student | balance | income | knn_probs | logistic_probs |
|---|---|---|---|---|---|
| No | No | 729.5265 | 44361.63 | 0 | 0.0012842 |
| No | Yes | 808.6675 | 17600.45 | 0 | 0.0019883 |
| No | Yes | 1220.5838 | 13268.56 | 0 | 0.0190870 |
| No | No | 237.0451 | 28251.70 | 0 | 0.0000843 |
| No | No | 606.7423 | 44994.56 | 0 | 0.0006514 |
| No | No | 286.2326 | 45042.41 | 0 | 0.0001107 |
Using a threshold
- Step 1: Predict probabilities for all observations
- Step 2: Set a threshold to obtain class labels (0.5 below)
threshold <- 0.5 # set threshold
default_test_wprobs <- default_test_wprobs |>
mutate(knn_preds = as_factor(if_else(knn_probs > threshold, "Yes", "No")),
logistic_preds = as_factor(if_else(logistic_probs > threshold, "Yes", "No"))
)
default_test_wprobs |> head() |> kable()| default | student | balance | income | knn_probs | logistic_probs | knn_preds | logistic_preds |
|---|---|---|---|---|---|---|---|
| No | No | 729.5265 | 44361.63 | 0 | 0.0012842 | No | No |
| No | Yes | 808.6675 | 17600.45 | 0 | 0.0019883 | No | No |
| No | Yes | 1220.5838 | 13268.56 | 0 | 0.0190870 | No | No |
| No | No | 237.0451 | 28251.70 | 0 | 0.0000843 | No | No |
| No | No | 606.7423 | 44994.56 | 0 | 0.0006514 | No | No |
| No | No | 286.2326 | 45042.41 | 0 | 0.0001107 | No | No |
Using a threshold
- Step 1: Predict probabilities for all observations
- Step 2: Set a threshold to obtain class labels (0.5 below)
threshold <- 0.5 # set threshold
default_test_wprobs <- default_test_wprobs |>
mutate(knn_preds = as_factor(if_else(knn_probs > threshold, "Yes", "No")),
logistic_preds = as_factor(if_else(logistic_probs > threshold, "Yes", "No")))
default_test_wprobs |> head() |> kable()| default | student | balance | income | knn_probs | logistic_probs | knn_preds | logistic_preds |
|---|---|---|---|---|---|---|---|
| No | No | 729.5265 | 44361.63 | 0 | 0.0012842 | No | No |
| No | Yes | 808.6675 | 17600.45 | 0 | 0.0019883 | No | No |
| No | Yes | 1220.5838 | 13268.56 | 0 | 0.0190870 | No | No |
| No | No | 237.0451 | 28251.70 | 0 | 0.0000843 | No | No |
| No | No | 606.7423 | 44994.56 | 0 | 0.0006514 | No | No |
| No | No | 286.2326 | 45042.41 | 0 | 0.0001107 | No | No |
Performance
Low Threshold
threshold <- 0.1 # set threshold
default_test_wprobs <- default_test_wprobs |>
mutate(knn_preds = as_factor(if_else(knn_probs > threshold, "Yes", "No")))
roc_metrics(default_test_wprobs, truth = default, estimate = knn_preds, event_level = "second") |> kable()| .metric | .estimator | .estimate |
|---|---|---|
| accuracy | binary | 0.9060000 |
| sensitivity | binary | 0.7364341 |
| specificity | binary | 0.9116507 |
High Threshold
threshold <- 0.9 # set threshold
default_test_wprobs <- default_test_wprobs |>
mutate(knn_preds = as_factor(if_else(knn_probs > threshold, "Yes", "No"))
)
roc_metrics(default_test_wprobs, truth = default, estimate = knn_preds, event_level = "second") |> kable()| .metric | .estimator | .estimate |
|---|---|---|
| accuracy | binary | 0.9685000 |
| sensitivity | binary | 0.0310078 |
| specificity | binary | 0.9997417 |
Question
- If I want to improve Recall/Sensitivity should I increase or decrease my threshold?
- If I want to improve my Precision/PPV should I increase or decrease my threshold?
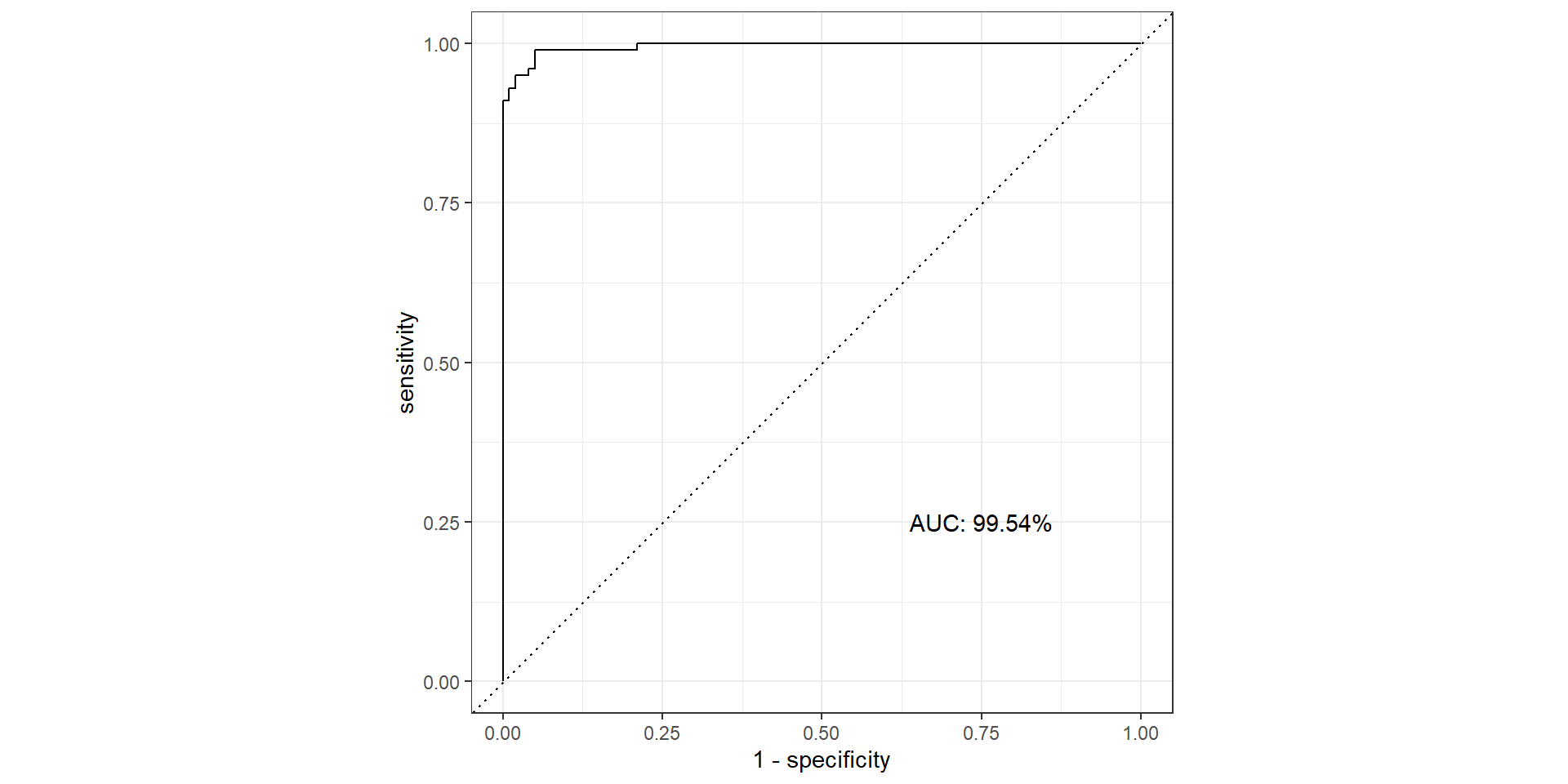
ROC Curve
ROC Curve and AUC
- ROC (Receiver Operating Characteristics) curve: popular graphic for comparing different classifiers across all possible thresholds
- Plots the (1-Specificity) along the x-axis and the Sensitivity (true positive rate) along the y-axis
- AUC: area under the AUC curve
- Ideal ROC curve will hug the top left corner
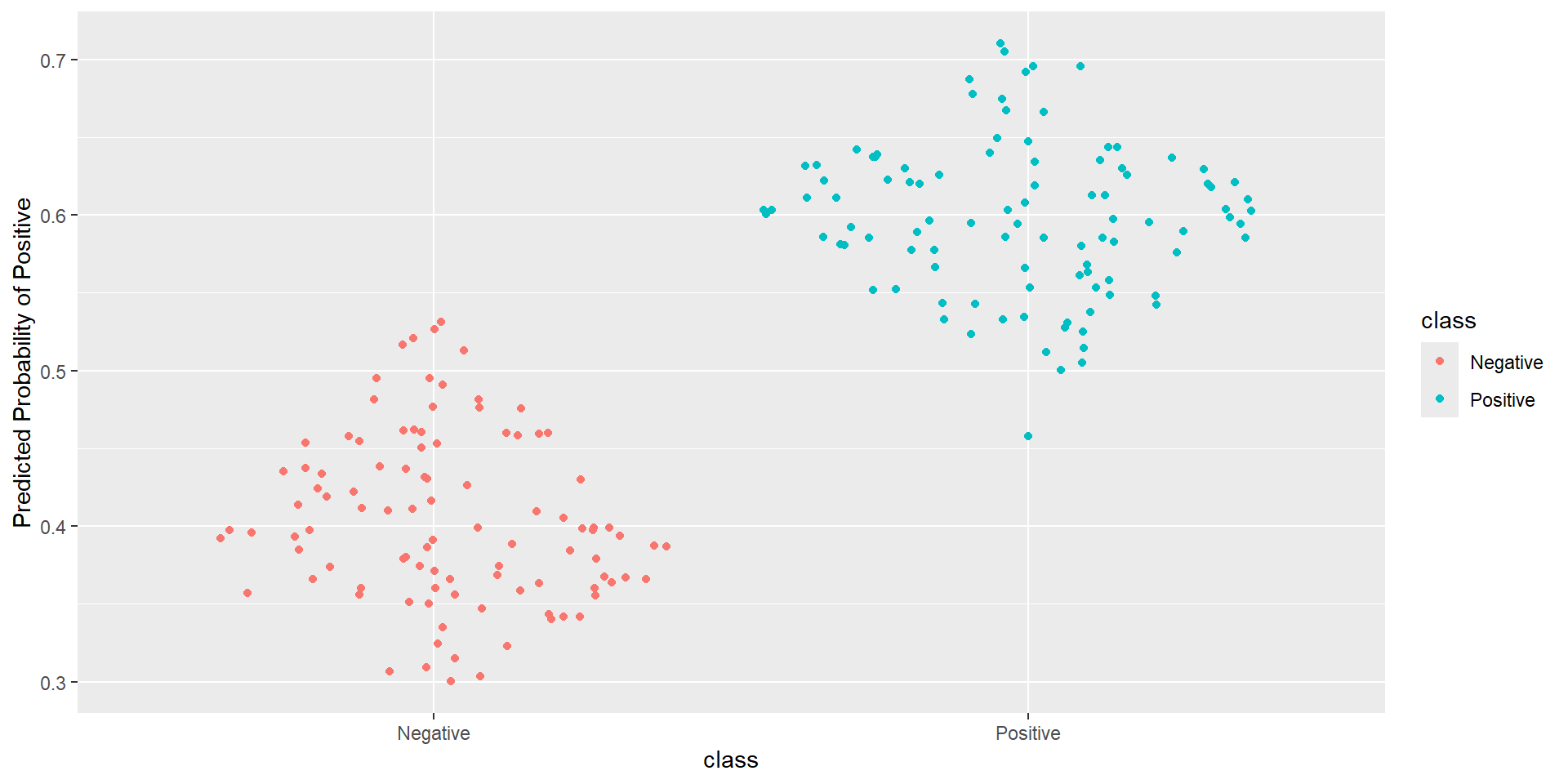
- Idea: How well is my classifier separating positives from negatives
ROC Curve
ROC Curve: Plot

AUC
- AUC: Area under the curve (ROC Curve that is)
- Measures how good your model is at separating categories
- Only for binary classification
AUC in R
Pathological Example 1
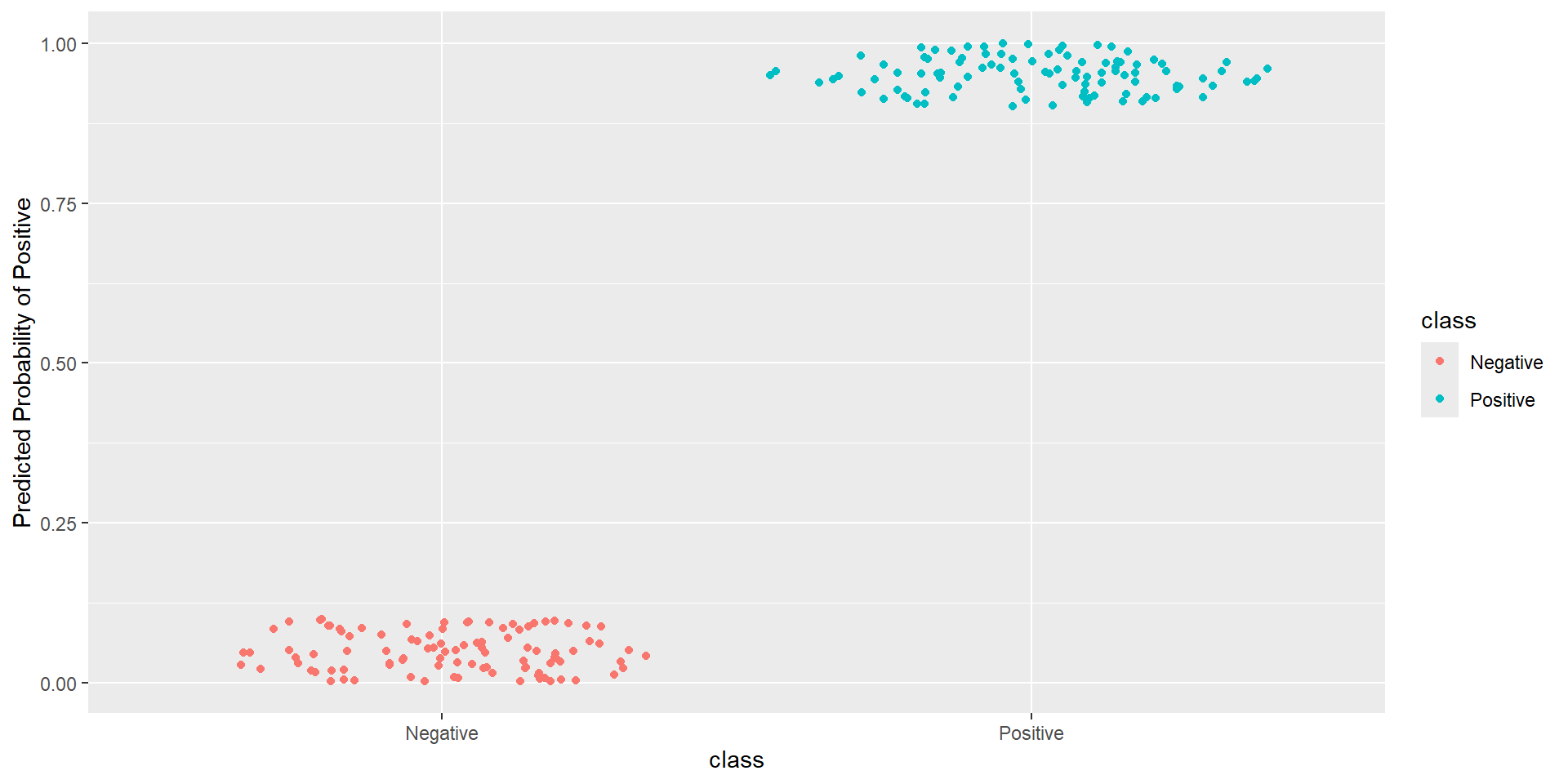
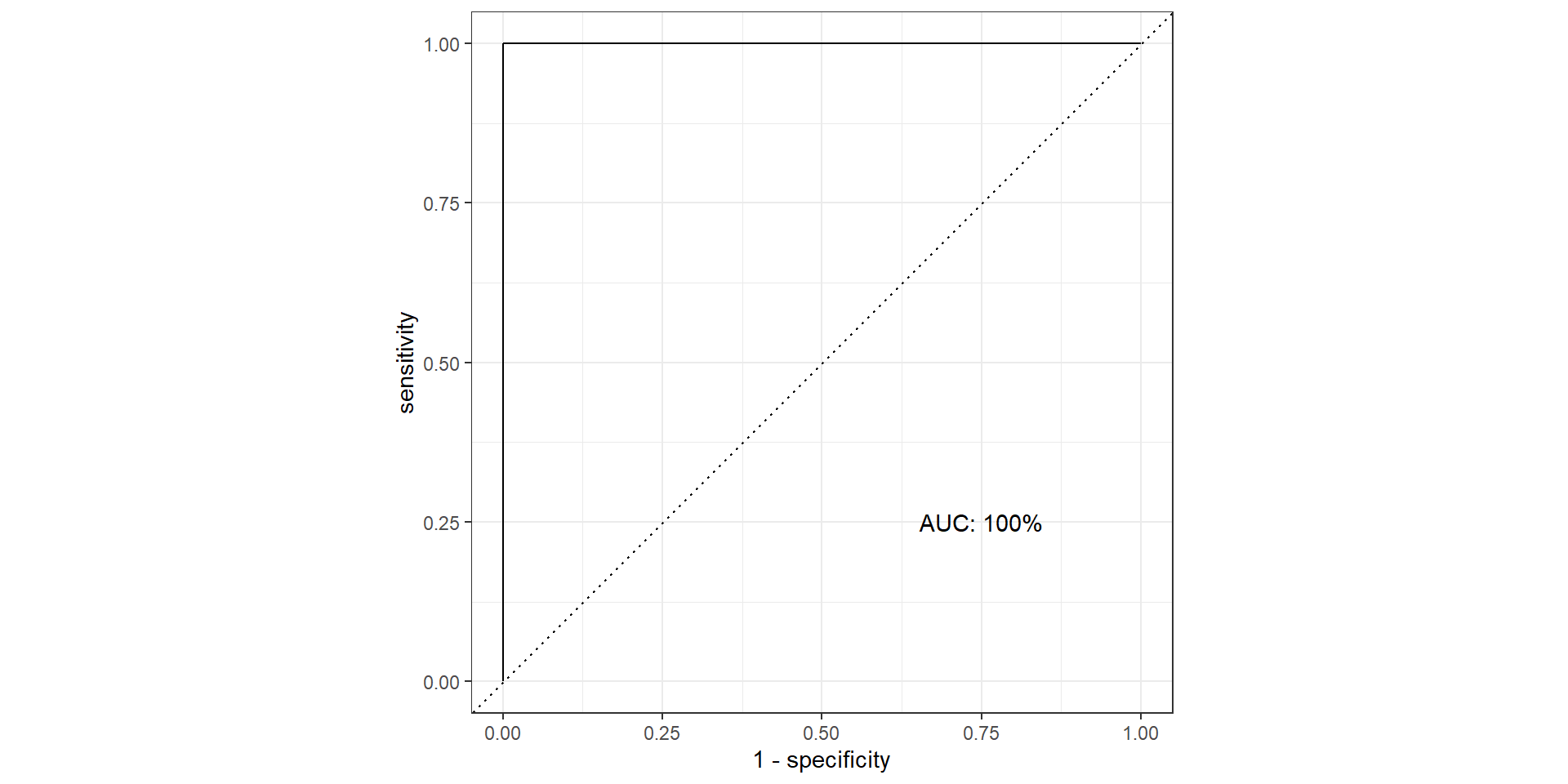
Pathological Example 2
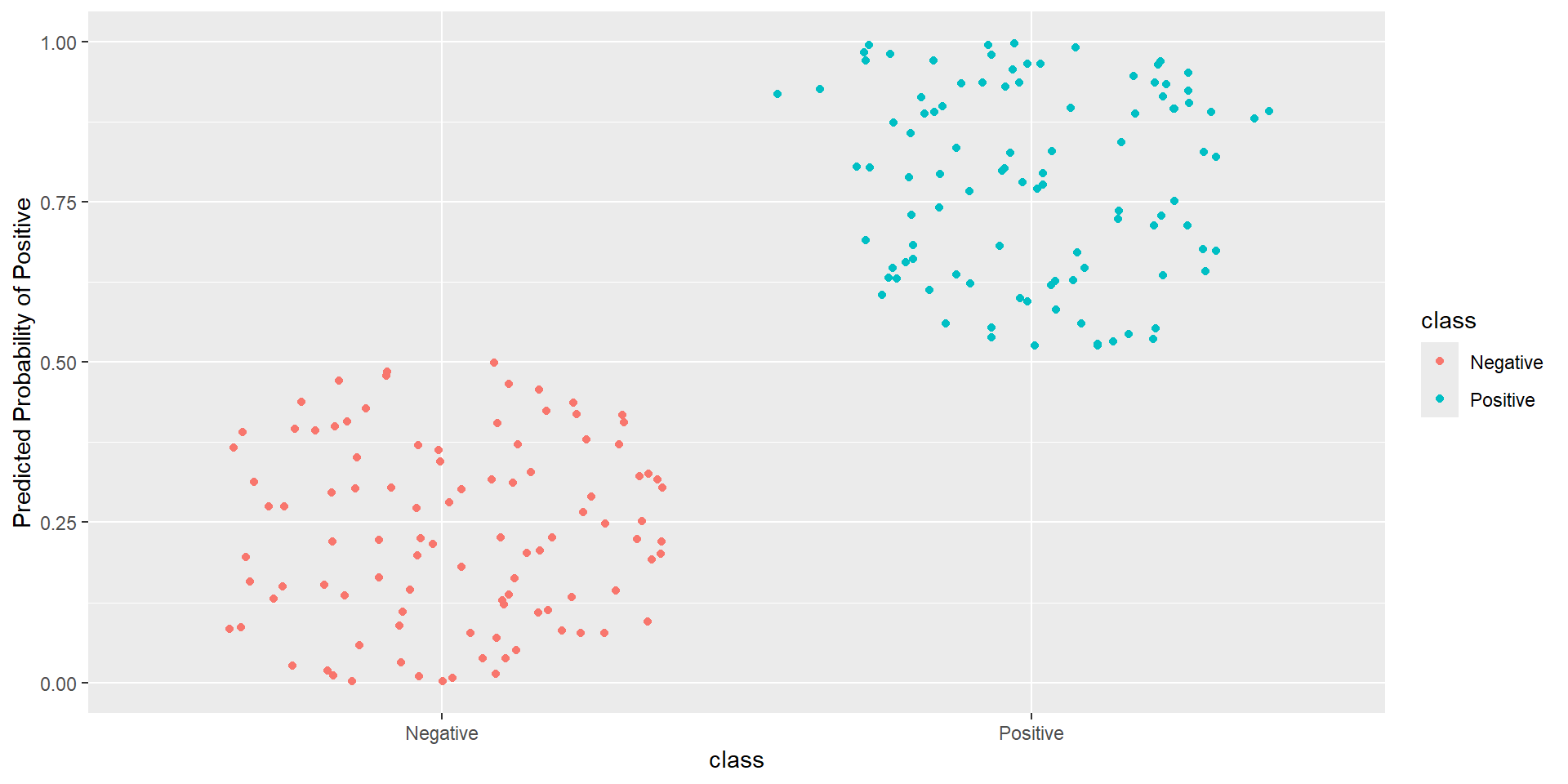
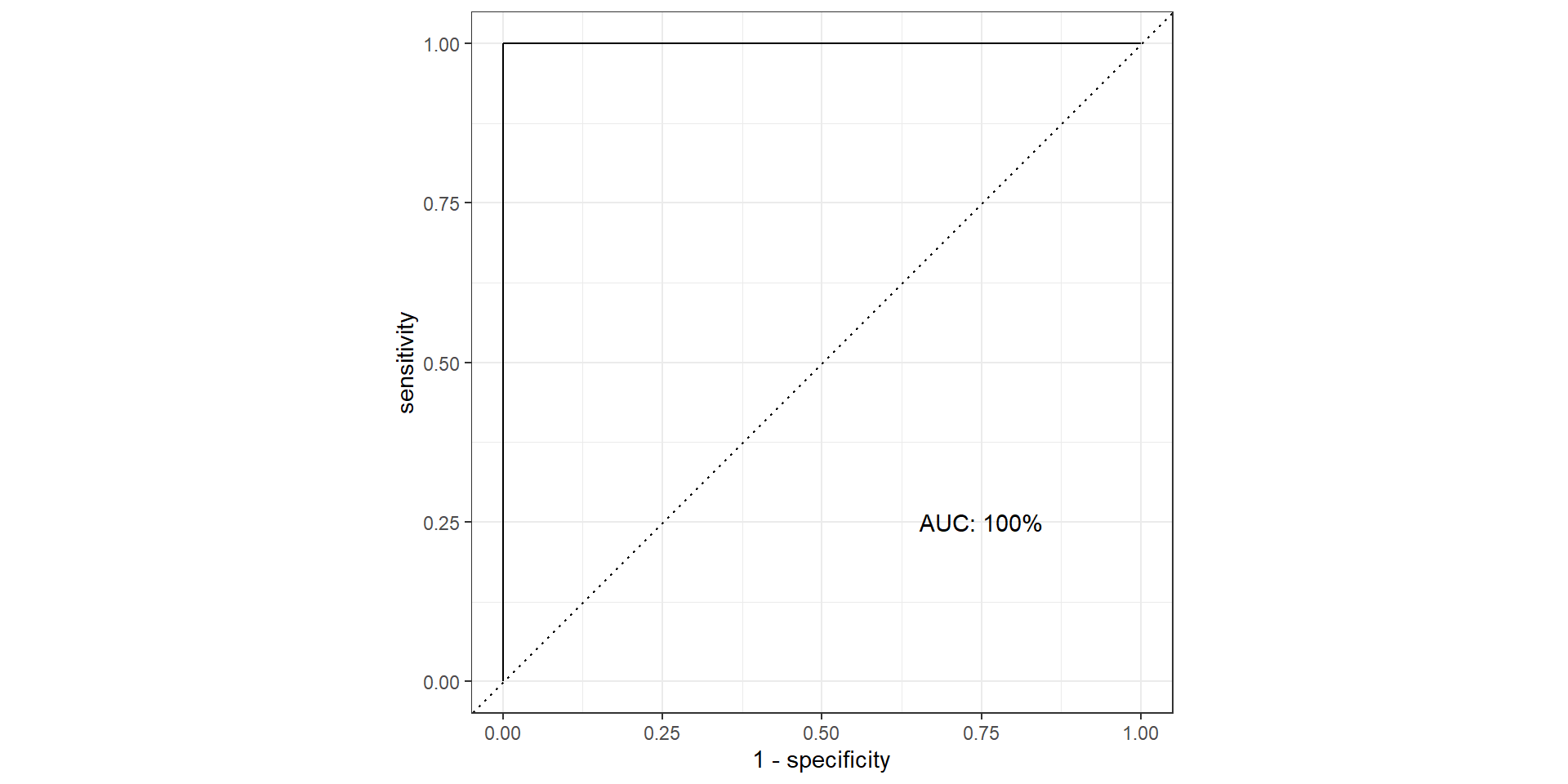
Pathological Example 3
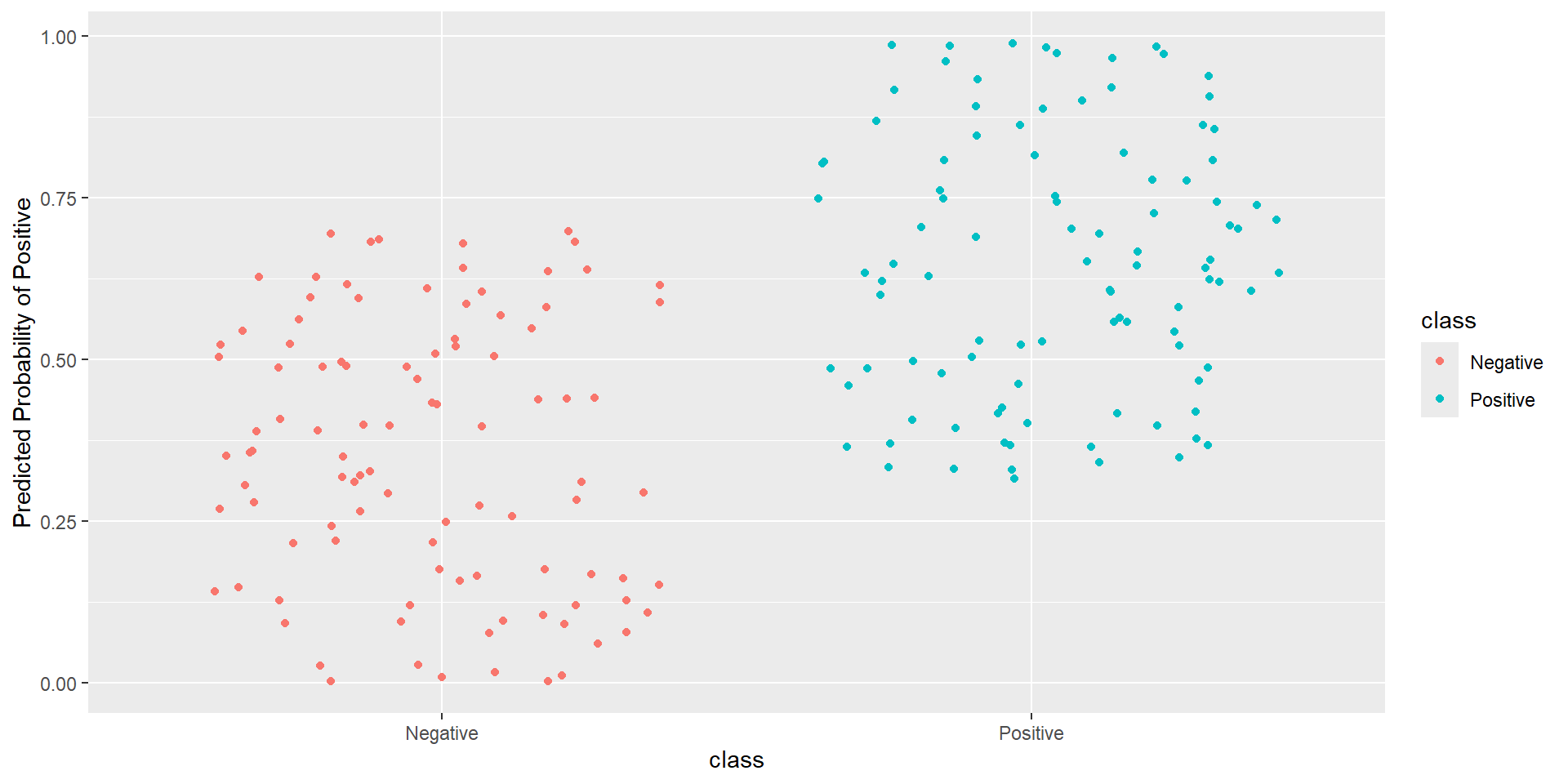
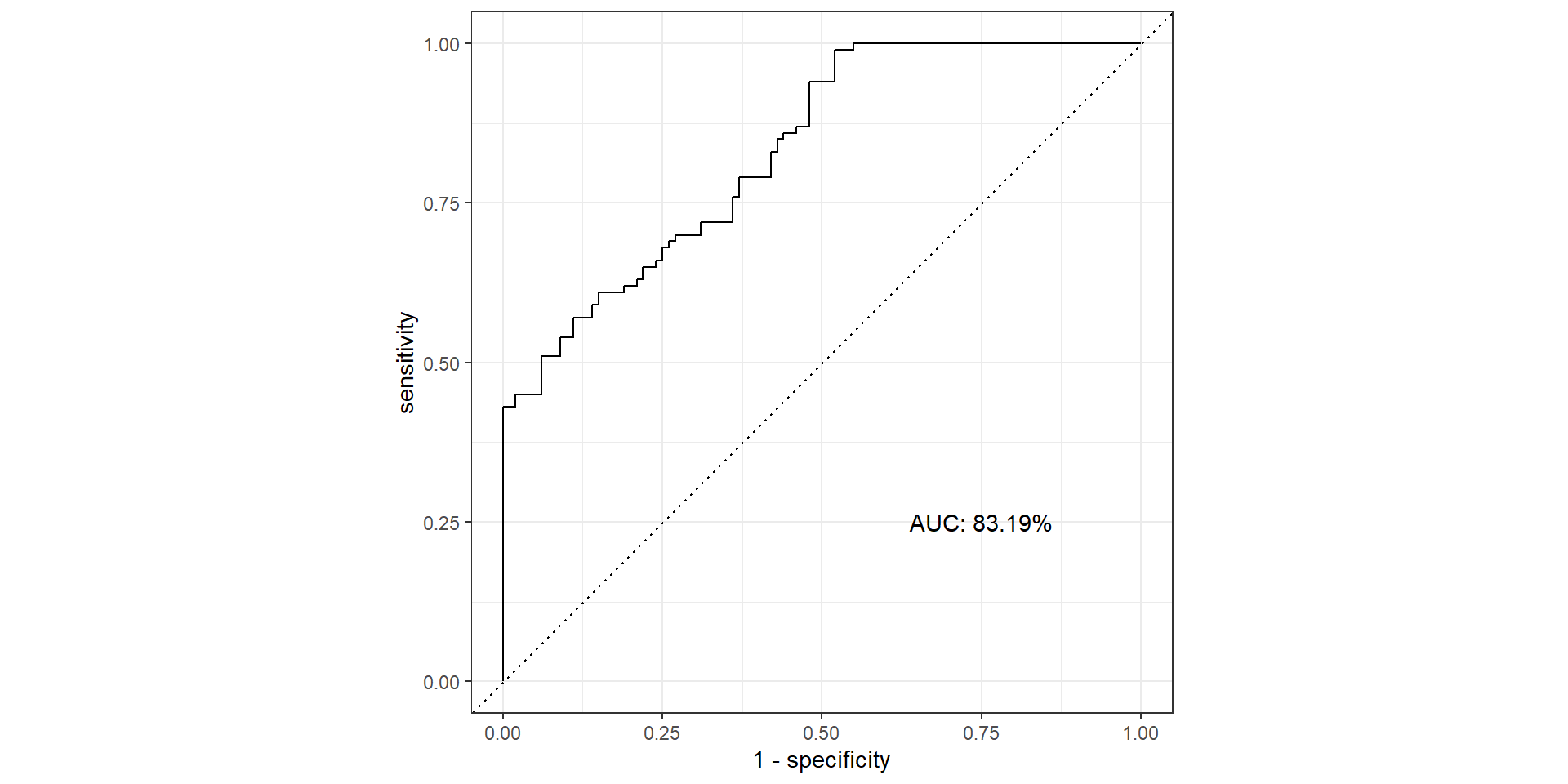
Pathological Example 4
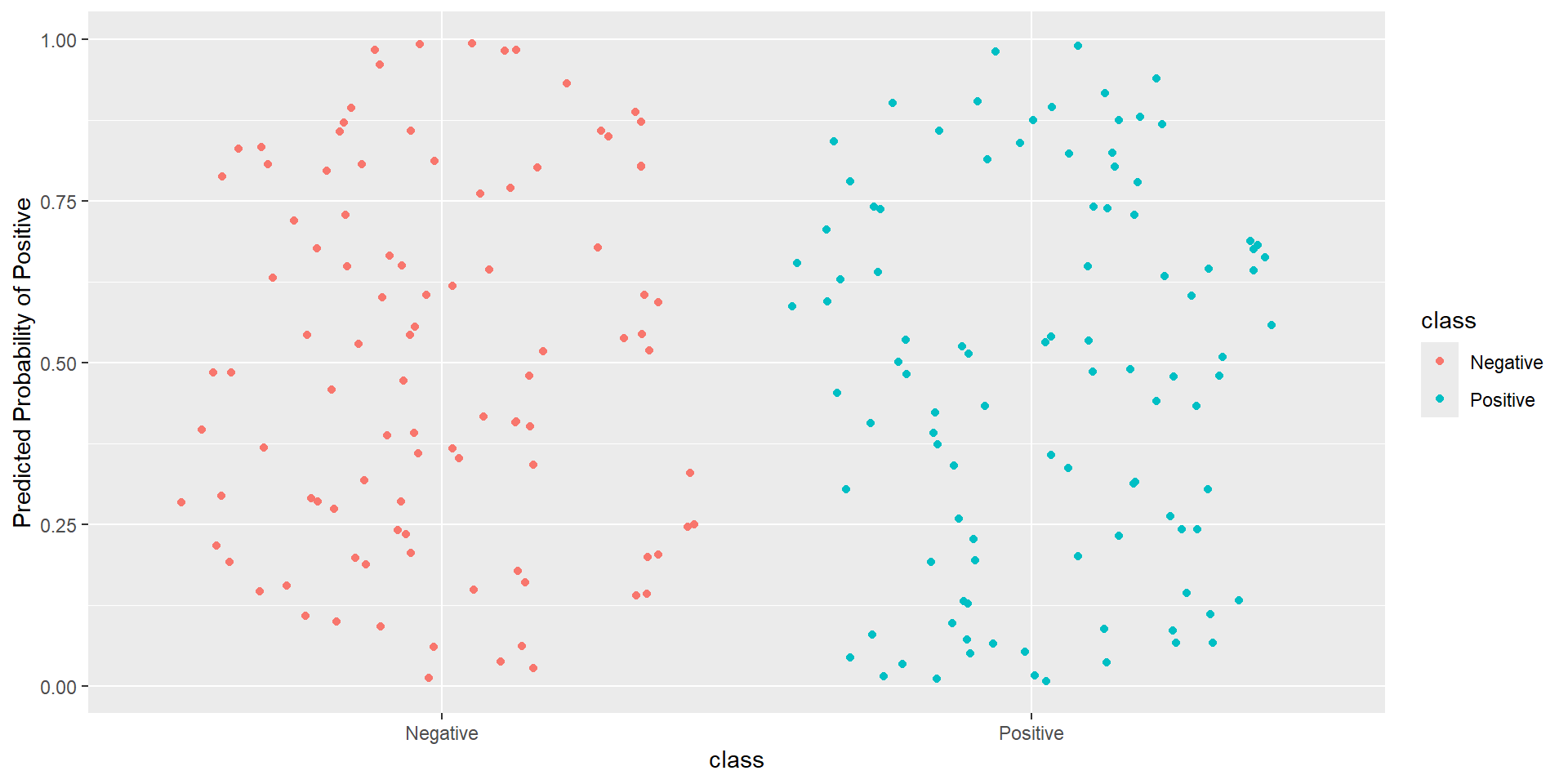
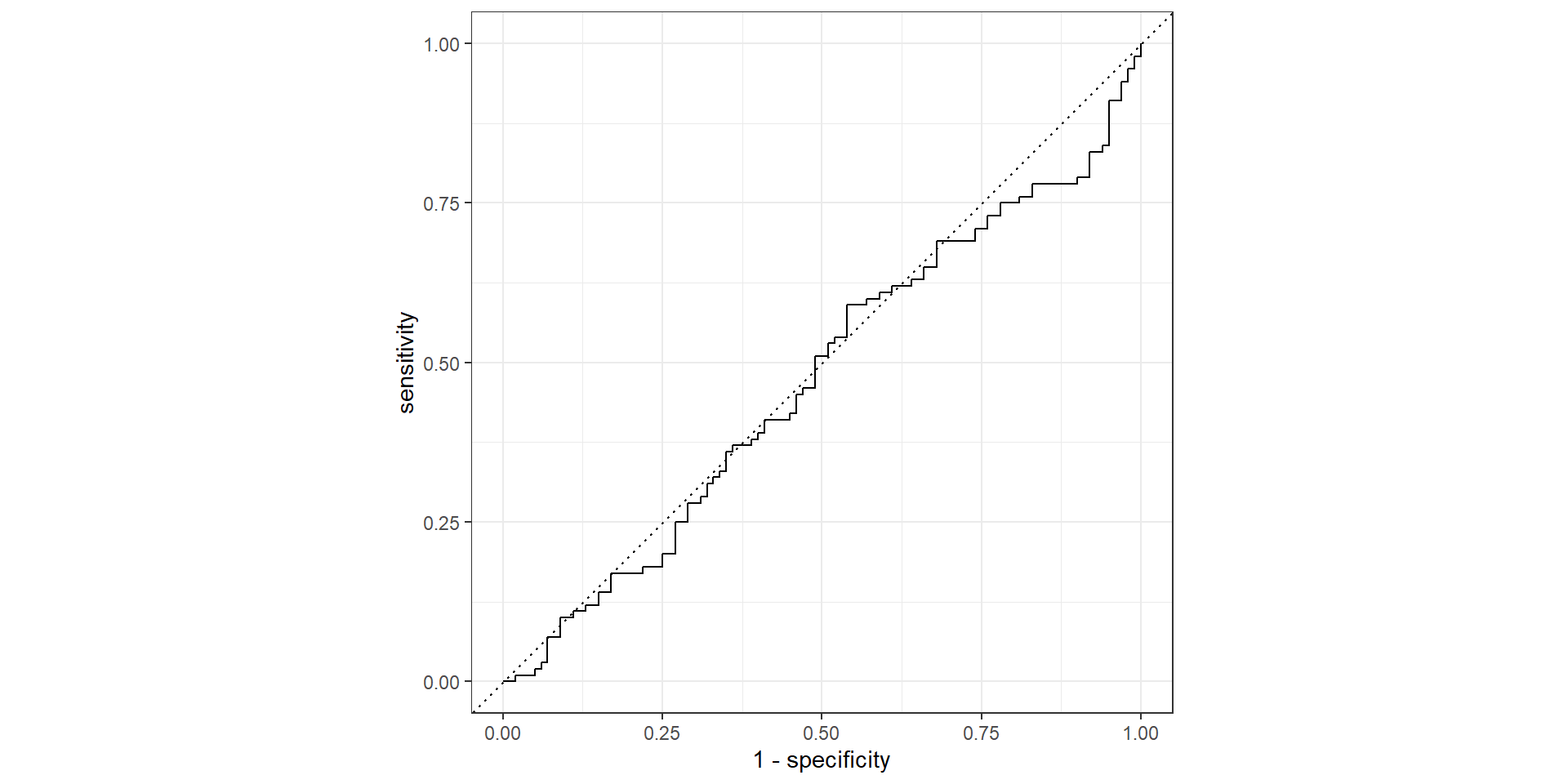
Pathological Example 5
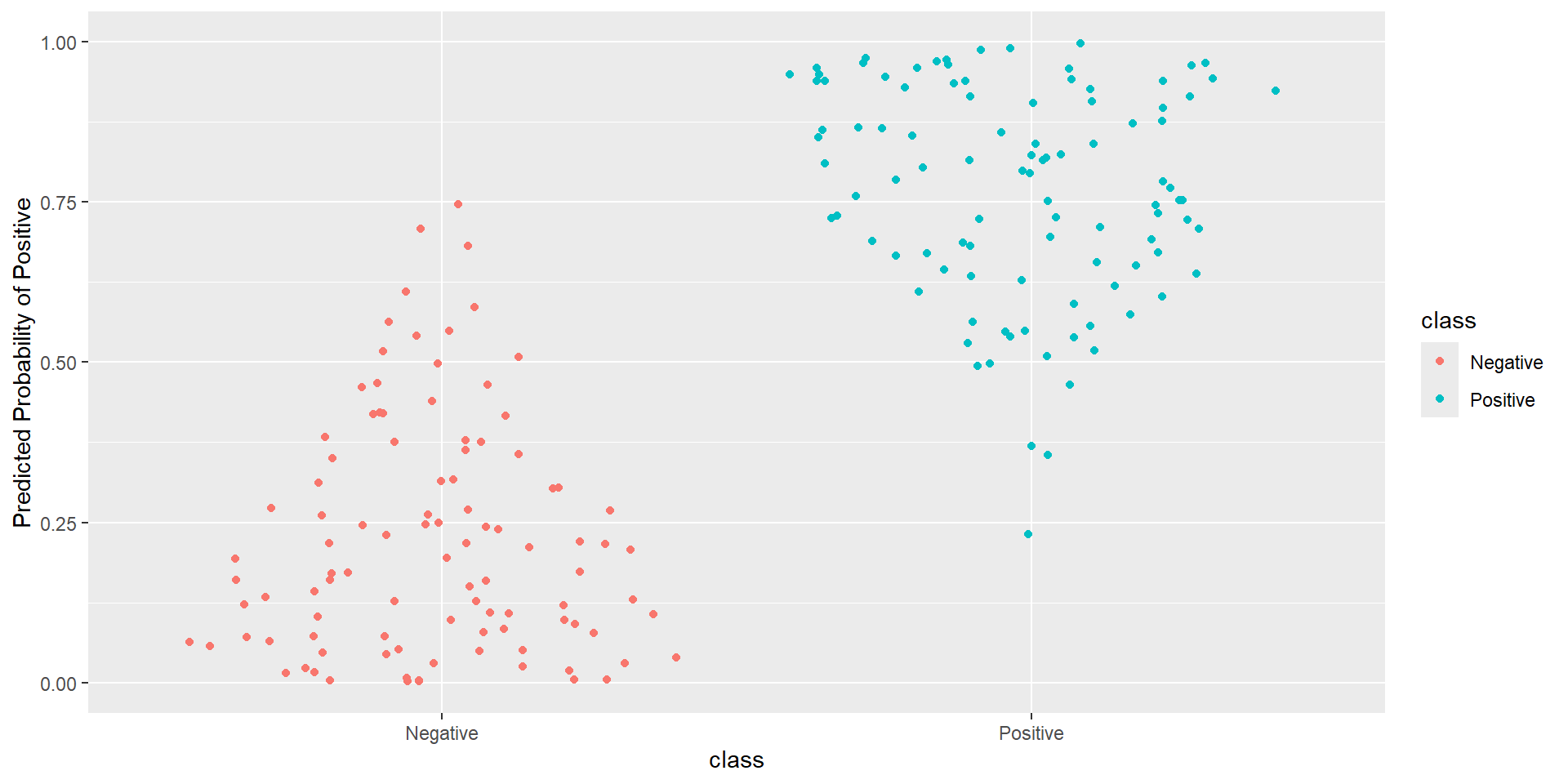
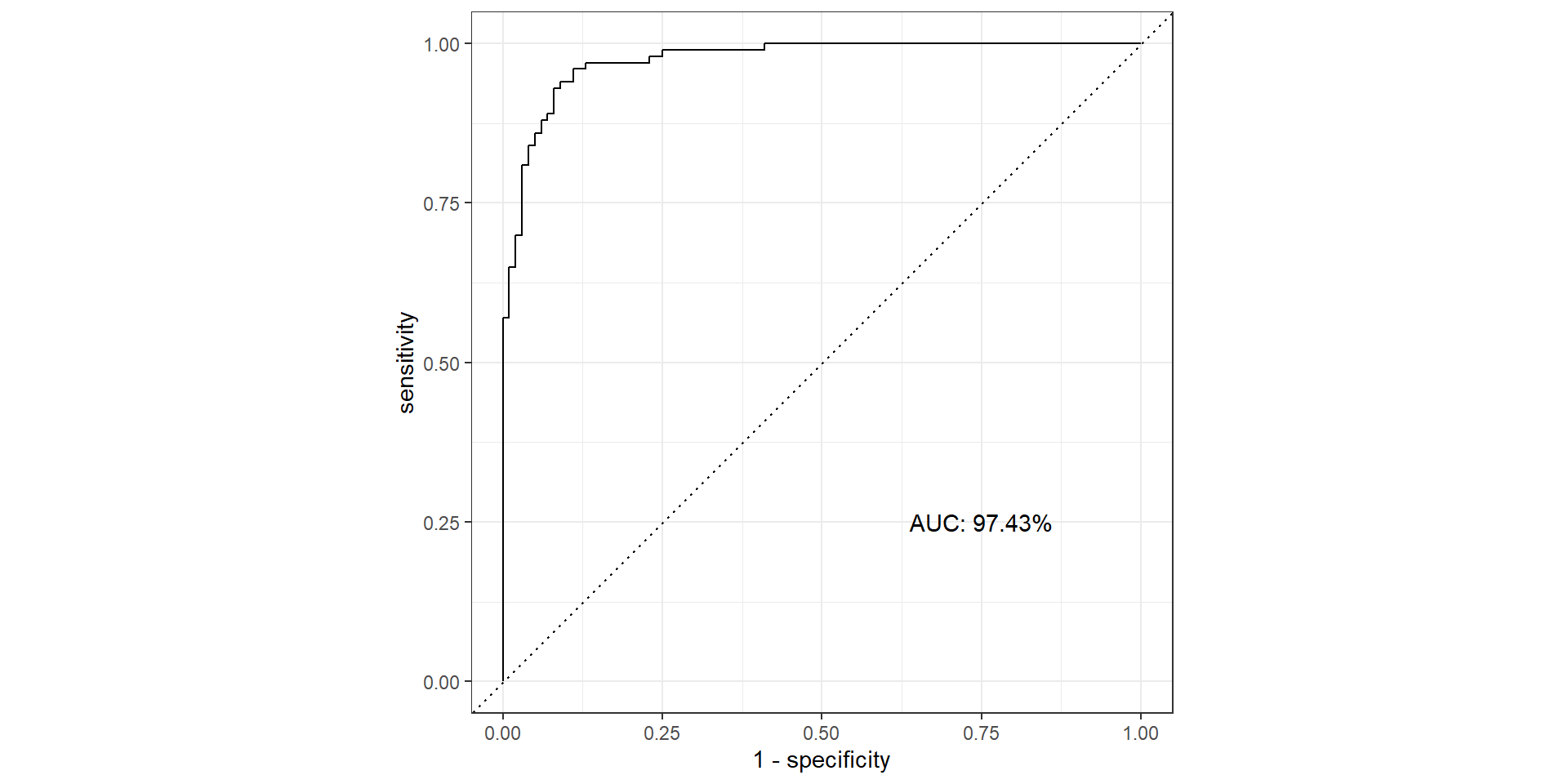
Pathological Example 6
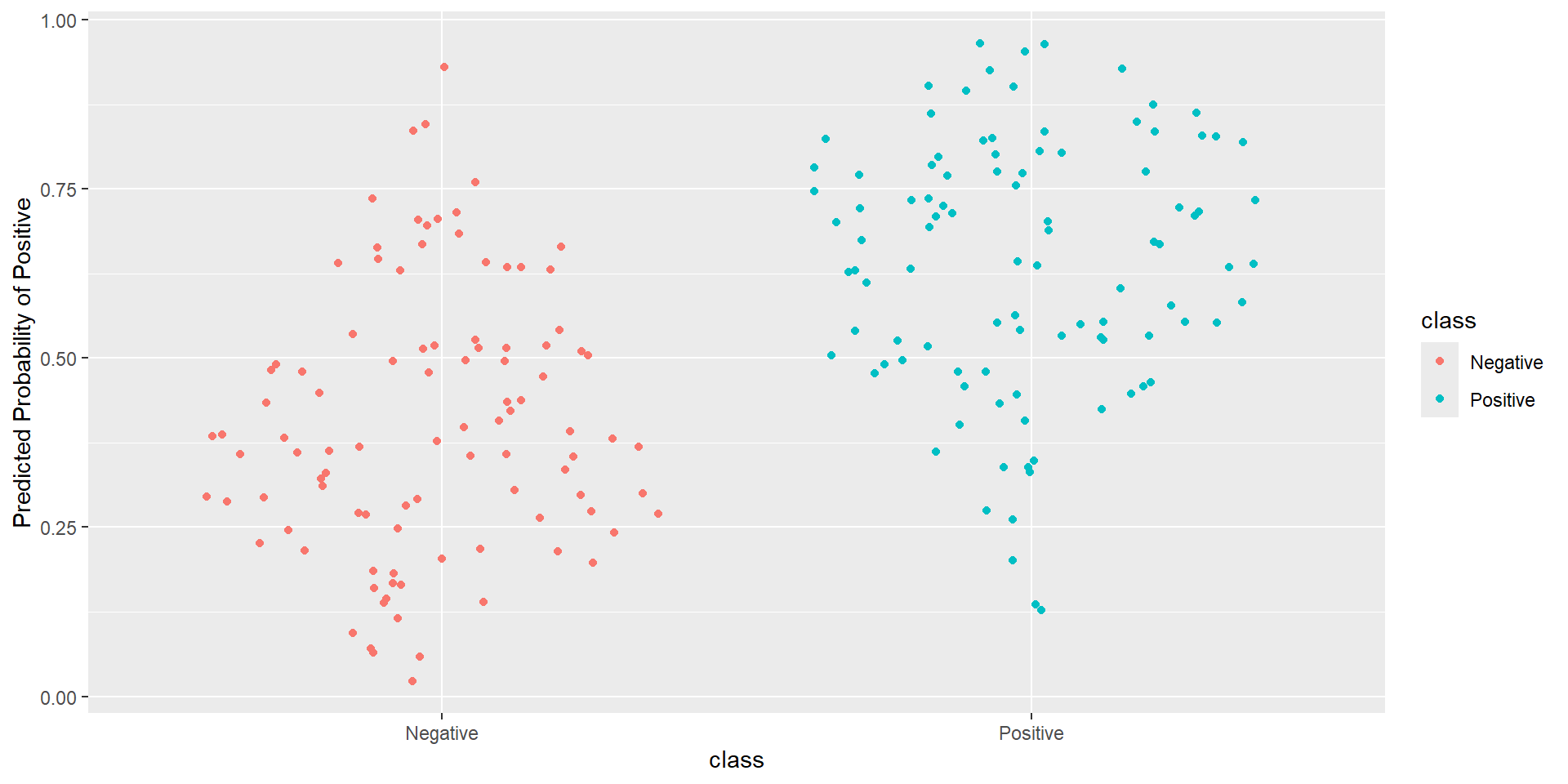
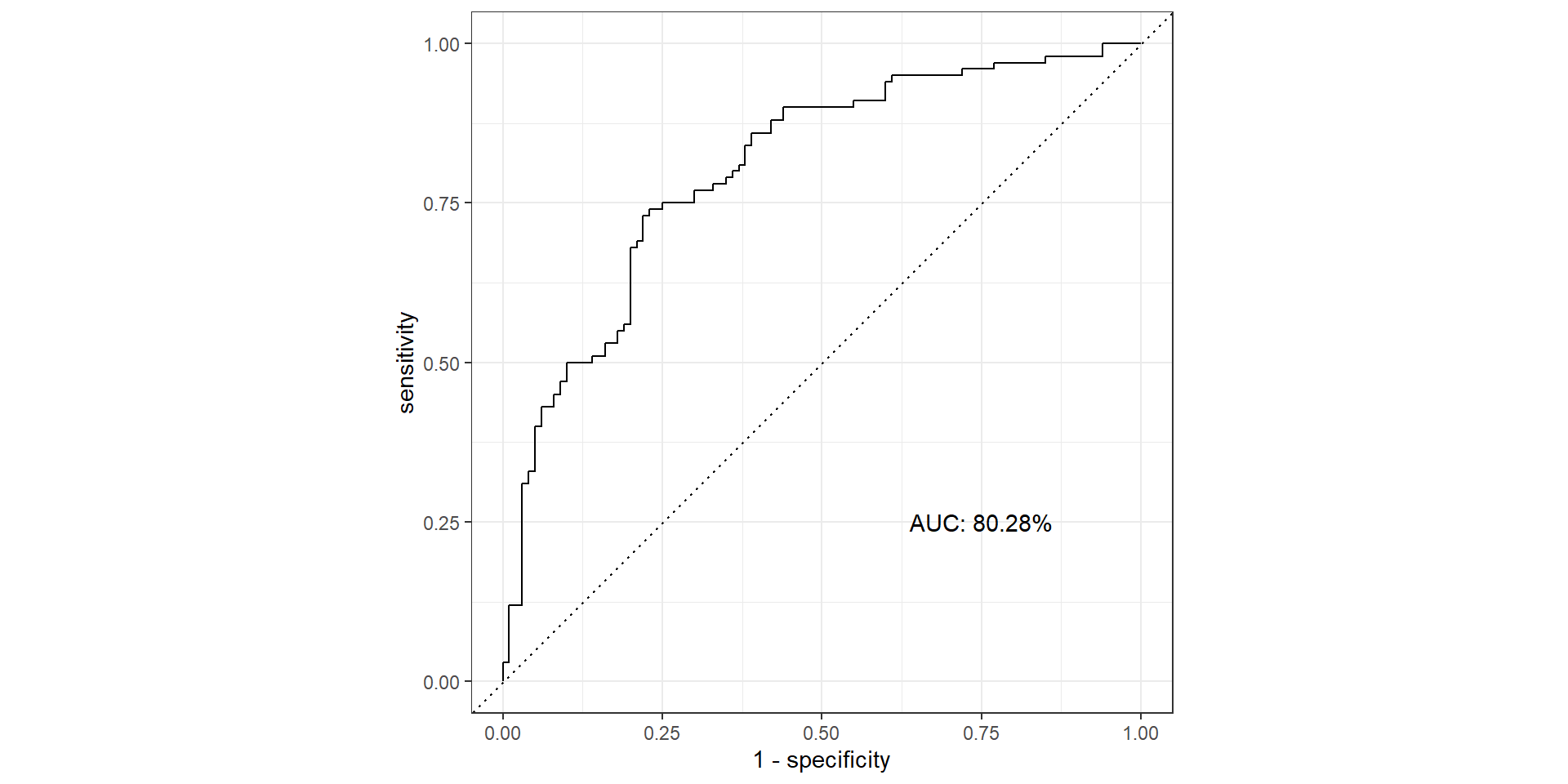
Pathological Example 7
AUC Questions
- What should be the minimum AUC?
- What should be that maximum possible AUC?
